Python-based CyTOF processing and analyzing package
Published:
CyTOF (Cytometry by time-of-flight) mass cytometry uses a time-of-flight ICP-MS (Inductively Coupled Plasma Mass Spectrometry) instrument (The Hyperion Imaging System (Fluidigm)) that can detect dozens of protein markers (up to 40) simultaneously. CyTOF mass cytometry is used to quantify labeled targets on the surface and interior of single cells by measuring the abundance of rare earth metal isotopes tagged to antibodies. Analyzing metal abundances enables the determination of marker expression in individual cells.
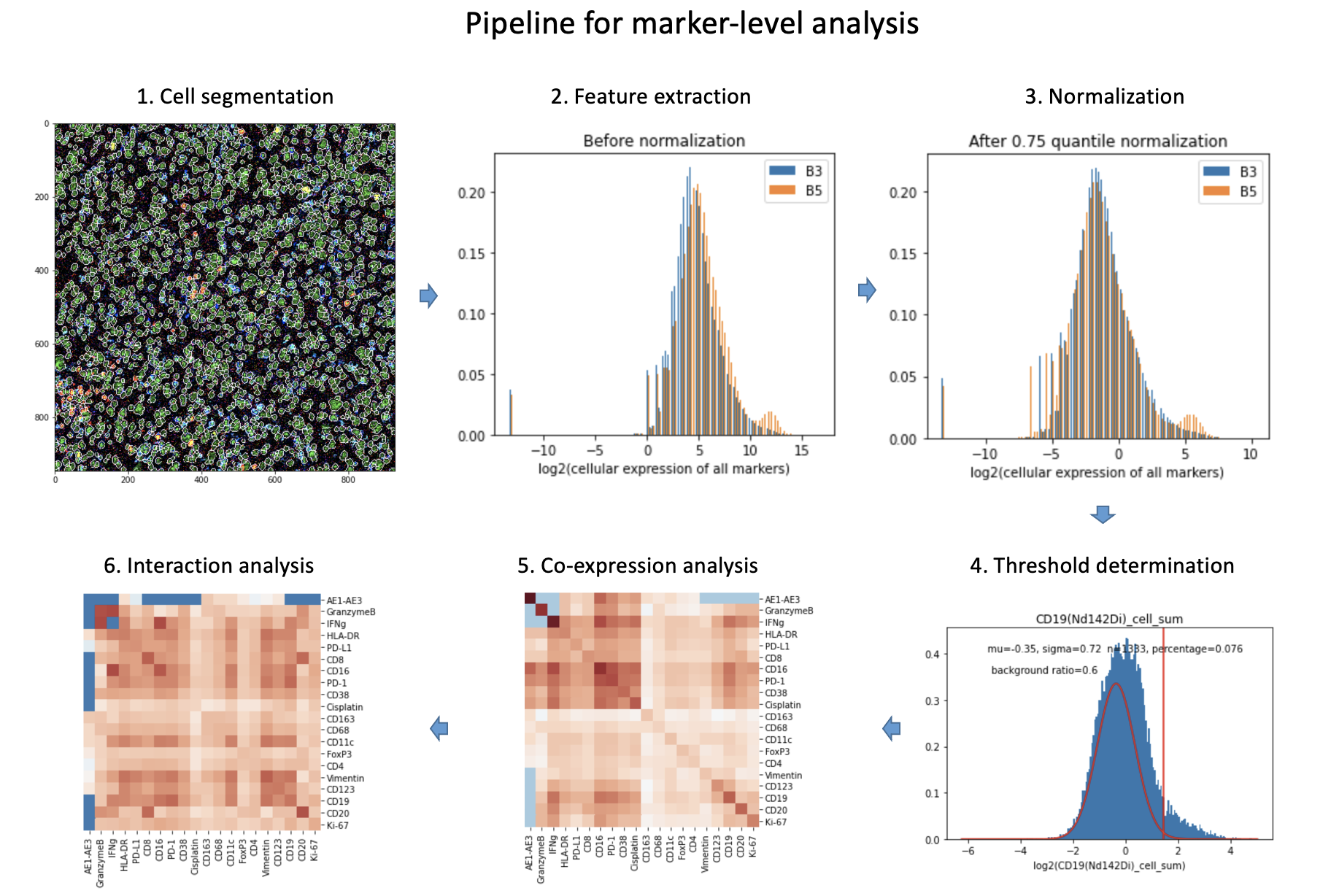
This python-based CyTOF image analysis package can be used as a stand-alone toolbox that is capable of automatically analysing CyTOF images. Main functionalities include preprocessing raw CyTOF data, visualization, nuclei and cell segmentation, single-cell feature extraction, exploration of individual cell phenotypes, cell-cell interactions, etc. Typically, data collected from the hyperion imaging system for one whole slide image (WSI) contains a MCD file (need MCD viewer to open) and several TXT files correspond to selected region-of-interests (ROIs) in the WSI. With this CyTOF image analysis package, no MCD Viewer or other tools are needed to be able to analyze CyTOF images.
Read more